Introduction to Logistic Regression
In this exercise, you will explore the characteristics of the logistic classifier.
The figure below shows a schematic of a multivariate logistic regression classifier.
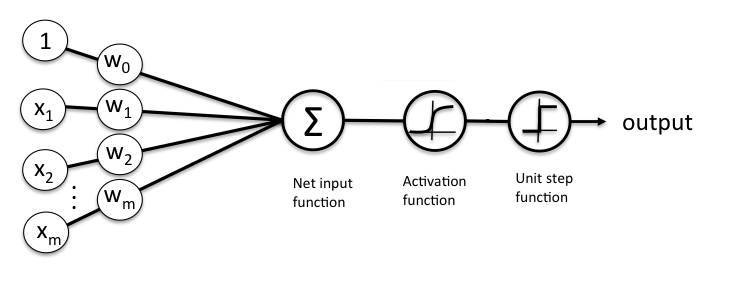
A Logistic regression classifier consists of 3 distinct parts:
A multivariate linear function : $z = \mathbf{w}^\top\mathbf{x} = \sum_i w_i*x_i +w_0$ , where $\mathbf{w}$ are the model parameters (including the bias term $w_0$) and $\mathbf{x}$ the homogeneous coordinates of the input
The sigmoid activation function $\sigma(z)=\frac{1}{1+\text{e}^{(-z)}}$
A binary step function (thresholding) at $\sigma(z)>0.5$.
The libraries used are imported in the next cell.
import numpy as np
import matplotlib.pyplot as pltExperimenting with model parameters of the logistic (regression) classifier
def linear_sigmoid(x,b,w) : # sigmoid function
"""
:param x: 1D array of the (single) input-feature values.
:param b: The bias parameter of the model.
:param W: The weight parameter of the model.
:return (float): output values of the sigmoid function.
"""
...
xs = np.linspace(-100,100,1000) ### linspace of "input features"
params = [3.0,0.15] ### parameters of the model
# Calc decision boundary here
Decision_boundary = ... # write your solution here
### plotting ####
plt.figure(figsize=(8,6))
plt.plot(xs, linear_sigmoid(xs,params[0],params[1]),label = r"$\sigma(x)$ = 1/(1 + exp(-(%.2f + %.2fx))"%(params[0],params[1]))
plt.plot([Decision_boundary,Decision_boundary],[0,1],'--C3',label = 'decision boundary')
plt.title(r"Sigmoid $\sigma(x)$", fontsize=20)
plt.legend()
plt.show()The cell below generates two classes (class1
and class2
) of data, drawn from normal distributions (with a different mean and variance)
### 2 classes of randomly generated data
x1= np.random.normal(-50,20,200)
y1 = np.zeros_like(x1)
x2= np.random.normal(50,30,200)
y2 = np.ones_like(x2)x1
and x2
are the variables for the input features of class1
and class2
, respectively. Likewise, y1
and y2
are the targets of class1
and class2
, respectively.
The following cell visualizes the data of the classes together with the sigmoid
values and the decision boundary.
plt.figure(figsize=(8,6))
plt.plot(xs, linear_sigmoid(xs,params[0],params[1]),label = r"$\sigma(x)$ = 1/(1 + exp(-(%.2f + %.2fx))"%(params[0],params[1]))
plt.plot(x1, y1+np.random.normal(0,0.01,200),'p',label ='class1')
plt.plot(x2, y2+np.random.normal(-0,0.01,200),'o',label = 'class2')
plt.plot([-params[0]/params[1],-params[0]/params[1]],[0,1],'--C3',label = 'decision boundary')
plt.title(r"Logistic regression and target data", fontsize=20)
plt.legend()
plt.show()def predict(x,w):
"""
:param x: 1D array of the (single) input-feature values.
:param w: The list of the model parameters, [bias, weight].
:return: Boolean array same size as x, where a True values signifies class2, and False signifies class1
"""
...The cell below visualizes the predictions.
x_all = np.concatenate([x1,x2])
y_all = np.concatenate([y1+np.random.normal(0,0.01,200),y2+np.random.normal(-0,0.01,200)])
y_bool = predict(x_all,params)
plt.figure(figsize=(8,6))
plt.plot(x_all[~y_bool], y_all[~y_bool],'p',color='orange',label ='Predicted class1')
plt.plot(x_all[y_bool], y_all[y_bool],'go',label = 'Predicted class2')
plt.plot(xs, linear_sigmoid(xs,params[0],params[1]),'b',label = r"$\sigma(x)$ = 1/(1 + exp(-(%.2f + %.2fx))"%(params[0],params[1]))
plt.plot([-params[0]/params[1],-params[0]/params[1]],[-0.04,1.04],'--C3',label = 'decision boundary')
plt.title(r"Logistic regression prediction", fontsize=20)
plt.legend()
plt.show()# Code you solution hereAccuracy of the logistic regression classifier is: 0.963
def sigmoid2D(X,params) : # sigmoid function
"""
:param X: tuple of input features (x,y).
:param parametes: List of the model parameters.
:return: output values of the sigmoid function.
"""
# Write solutions here
...
#### Plotting
x = np.linspace(-50, 50, 1000)
y = np.linspace(-50, 50, 1000)
X, Y = np.meshgrid(x, y)
params3 = [0.07,0.037,0.085]
Z = sigmoid2D((X, Y),params3)
fig = plt.figure(figsize=(15,7))
# ----- First subplot: 3D sigmoid function -----
ax1 = fig.add_subplot(1, 2, 1, projection='3d') # 1 row, 2 columns, first plot
# Decision boundary plane calculation
y_decision_boundary = -(params3[0]/params3[2]) - (params3[1]/params3[2]) * x
# Create mesh for decision boundary plane in 3D space
Y_decision, Z_decision = np.meshgrid(y_decision_boundary, np.linspace(0, 1, 1000)) # z from 0 to 1 for probabilities
X_decision = np.tile(x, (len(Y_decision), 1)) # x values remain constant for the plane
# Plot the decision boundary as a wireframe to make it clearly visible
boundary_surface = ax1.plot_wireframe(X_decision, Y_decision, Z_decision, color='black', alpha=0.6, label='Decision Boundary')
sigmoid_surface = ax1.plot_surface(X, Y, Z, color='green', alpha=0.3) # making it more transparent
ax1.set_title(r"Sigmoid : $\sigma(x,y)$ = 1/(1 + exp(-(%.2f + %.2fx + %.2fy))"%(params3[0],params3[1],params3[2]), fontsize=20)
ax1.set_xlabel('X')
ax1.set_ylabel('Y')
ax1.set_zlabel('Z (Probability)')
ax1.view_init(elev=15., azim=2)
# ----- Second subplot: 2D projection of the decision boundary -----
ax2 = fig.add_subplot(1, 2, 2) # 1 row, 2 columns, second plot
# Plot the decision boundary line
ax2.plot(x, y_decision_boundary, color='black',linewidth=3, label='Decision Boundary')
ax2.fill_between(x, y_decision_boundary, y[0], color='blue', alpha=0.3, label = 'class 1') # Optional: fill area under the line
ax2.fill_between(x, y_decision_boundary, y[-1], color='red', alpha=0.3, label = 'class 2') # Optional: fill area under the line
ax2.set_xlabel('X')
ax2.set_ylabel('Y')
ax2.set_title('Decision Boundary in XY plane', fontsize=14)
ax2.legend()
plt.tight_layout()
plt.show()# write your reflections hereIdentically to above, we generate 2 classes (class1
and class2
), but this time with two input features.
S = 100*np.eye(2)
p1,q1 = np.random.multivariate_normal([20,8], S, 200).T
p2,q2 = np.random.multivariate_normal([-10,-13], S, 200).T
z1 = np.ones_like(q1)+np.random.normal(0,0.01,200)
z2 = np.zeros_like(q2)+np.random.normal(0,0.01,200)def predict2D(X,params):
"""
:param x: tuple of 1D arrays of the input-feature values.
:param params: The list of the model parameters, [bias, weight1, weight2].
:return: Boolean array same size as x, where a True values signifies class2, and False signifies class1
"""
...
# Code you solution hereAccuracy of the logistic regression classifier is: 0.037
# Fill the empty list below with the best model parameters from task 3
params3 = []
x = np.linspace(-50, 50, 1000)
y = np.linspace(-50, 50, 1000)
X, Y = np.meshgrid(x, y)
Z = sigmoid2D((X, Y), params3)
# Decision boundary plane calculation
y_decision_boundary = -(params3[0] / params3[2]) - (params3[1] / params3[2]) * x
# Separate plot for the second subplot
fig, ax2 = plt.subplots(figsize=(10, 7))
# Plot the decision boundary line
ax2.plot(x, y_decision_boundary, color='black', linewidth=3, label='Decision Boundary')
ax2.plot(p1, q1, 'p', color='blue', label='class 1')
ax2.plot(p2, q2, 'o', color='red', label='class 2')
ax2.fill_between(x, y_decision_boundary, y[0], color='red', alpha=0.3, label='class 2 predictions')
ax2.fill_between(x, y_decision_boundary, y[-1], color='blue', alpha=0.3, label='class 1 predictions')
ax2.set_xlabel('X')
ax2.set_ylabel('Y')
ax2.set_title('Decision Boundary in XY plane', fontsize=14)
ax2.legend()
plt.show()# Fill the empty list below with the best model parameters from task 3
params3 = []
# Plotting data point along with the sigmoid
x = np.linspace(-50, 50, 1000)
y = np.linspace(-50, 50, 1000)
X, Y = np.meshgrid(x, y)
Z = sigmoid2D((X, Y),params3)
fig = plt.figure(figsize=(15,7))
# ----- First subplot: 3D sigmoid function -----
ax1 = fig.add_subplot(1, 1, 1, projection='3d') # 1 row, 2 columns, first plot
# Decision boundary plane calculation
y_decision_boundary = -(params3[0]/params3[2]) - (params3[1]/params3[2]) * x
# Create mesh for decision boundary plane in 3D space
Y_decision, Z_decision = np.meshgrid(y_decision_boundary, np.linspace(0, 1, 1000)) # z from 0 to 1 for probabilities
X_decision = np.tile(x, (len(Y_decision), 1)) # x values remain constant for the plane
# Plot the decision boundary as a wireframe to make it clearly visible
boundary_surface = ax1.plot_wireframe(X_decision, Y_decision, Z_decision, color='black', alpha=0.6, label='Decision Boundary')
sigmoid_surface = ax1.plot_surface(X, Y, Z, color='green', alpha=0.3) # making it more transparent
ax1.scatter(p1,q1,z1,'p',color ='red',label='class 1')
ax1.scatter(p2,q2,z2,'o',color='blue', label='class 2')
ax1.set_title(r"Sigmoid : $\sigma(x,y)$ = 1/(1 + exp(-(%.2f + %.2fx + %.2fy))"%(params3[0],params3[1],params3[2]), fontsize=20)
ax1.set_xlabel('X')
ax1.set_ylabel('Y')
ax1.set_zlabel('Z (Probability)')
ax1.view_init(elev=15., azim=2)
plt.tight_layout()
plt.show()# write your reflections here